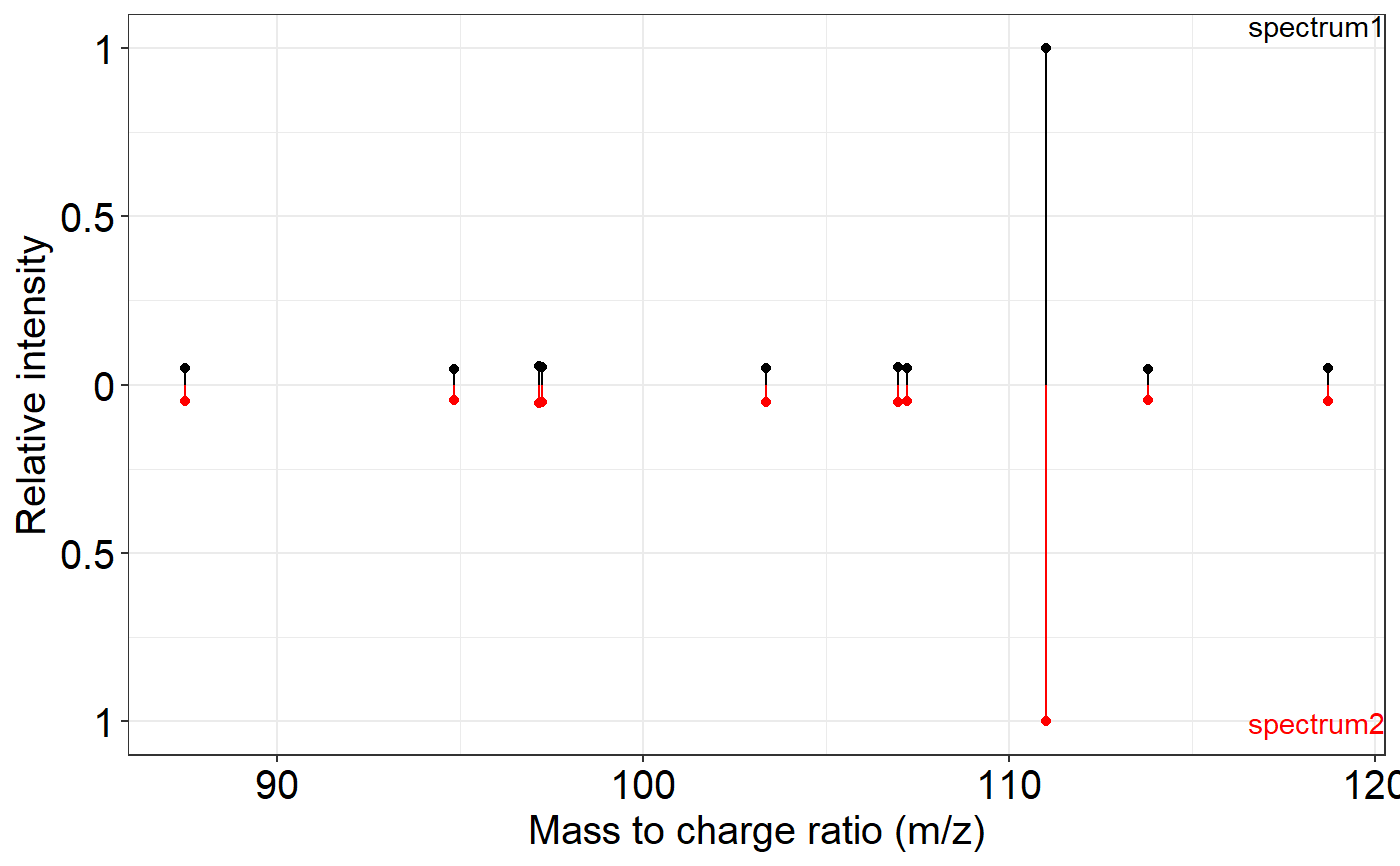
This function provides a method to visualize and compare two MS2 spectra side by side. The method to be used for the comparison depends on the class of the `spectrum1` and `spectrum2` objects provided.
ms2_plot(
spectrum1,
spectrum2,
spectrum1_name = "spectrum1",
spectrum2_name = "spectrum2",
range.mz,
ppm.tol = 30,
mz.ppm.thr = 400,
xlab = "Mass to charge ratio (m/z)",
ylab = "Relative intensity",
col1 = "red",
col2 = "black",
title.size = 15,
lab.size = 15,
axis.text.size = 15,
legend.title.size = 15,
legend.text.size = 15,
interactive_plot = FALSE
)
# S3 method for class 'data.frame'
ms2_plot(
spectrum1,
spectrum2,
spectrum1_name = "spectrum1",
spectrum2_name = "spectrum2",
range.mz,
ppm.tol = 30,
mz.ppm.thr = 400,
xlab = "Mass to charge ratio (m/z)",
ylab = "Relative intensity",
col1 = "red",
col2 = "black",
title.size = 15,
lab.size = 15,
axis.text.size = 15,
legend.title.size = 15,
legend.text.size = 15,
interactive_plot = FALSE
)
# S3 method for class 'matrix'
ms2_plot(
spectrum1,
spectrum2,
spectrum1_name = "spectrum1",
spectrum2_name = "spectrum2",
range.mz,
ppm.tol = 30,
mz.ppm.thr = 400,
xlab = "Mass to charge ratio (m/z)",
ylab = "Relative intensity",
col1 = "red",
col2 = "black",
title.size = 15,
lab.size = 15,
axis.text.size = 15,
legend.title.size = 15,
legend.text.size = 15,
interactive_plot = FALSE
)Arguments
- spectrum1
A spectrum object representing the first MS2 spectrum.
- spectrum2
A spectrum object representing the second MS2 spectrum.
- spectrum1_name
A character string specifying the name/label for the first spectrum. Default is "spectrum1".
- spectrum2_name
A character string specifying the name/label for the second spectrum. Default is "spectrum2".
- range.mz
Numeric vector of length 2 specifying the range of m/z values to display in the plot.
- ppm.tol
Numeric value specifying the ppm tolerance for matching peaks between the spectra. Default is 30.
- mz.ppm.thr
Numeric value specifying the threshold for m/z ppm difference. Default is 400.
- xlab
Character string specifying the x-axis label. Default is "Mass to charge ratio (m/z)".
- ylab
Character string specifying the y-axis label. Default is "Relative intensity".
- col1
Character string specifying the color for the first spectrum. Default is "red".
- col2
Character string specifying the color for the second spectrum. Default is "black".
- title.size
Numeric value specifying the font size for the plot title. Default is 15.
- lab.size
Numeric value specifying the font size for x and y labels. Default is 15.
- axis.text.size
Numeric value specifying the font size for axis text. Default is 15.
- legend.title.size
Numeric value specifying the font size for the legend title. Default is 15.
- legend.text.size
Numeric value specifying the font size for the legend text. Default is 15.
- interactive_plot
Logical indicating whether the plot should be interactive (e.g., using plotly). Default is FALSE.
Value
A plot visualizing the comparison between the two provided MS2 spectra.
See also
The specific plotting methods associated with different spectrum classes that implement this generic function.
Examples
# Assuming `spec1` and `spec2` are spectrum objects
spec1 <- data.frame(
mz = c(
87.50874,
94.85532,
97.17808,
97.25629,
103.36186,
106.96647,
107.21461,
111.00887,
113.79269,
118.70564
),
intensity =
c(
8356.306,
7654.128,
9456.207,
8837.188,
8560.228,
8746.359,
8379.361,
169741.797,
7953.080,
8378.066
)
)
spec2 <- spec1
ms2_plot(spec1, spec2)
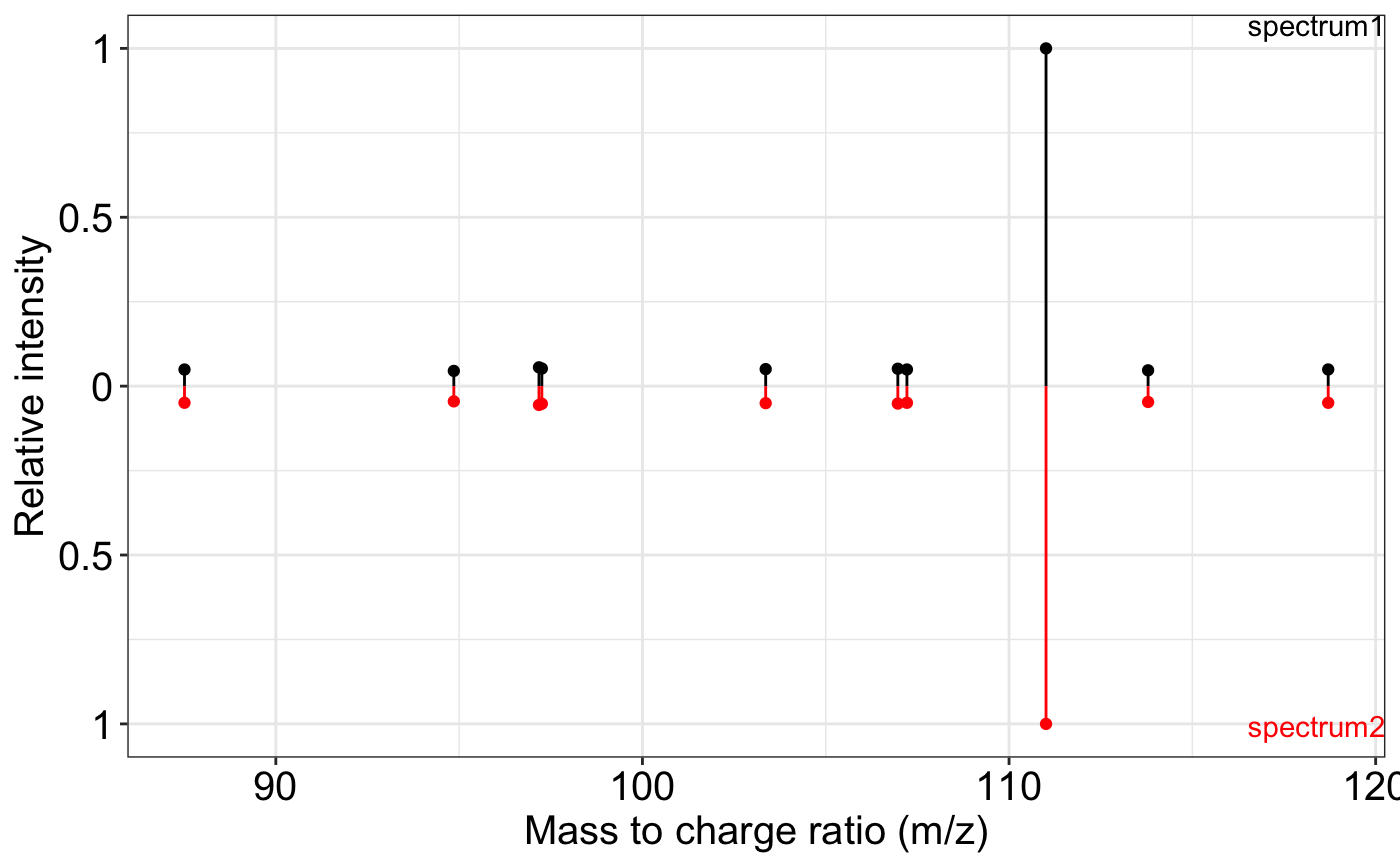 spectrum1 <- data.frame(
mz = c(
87.50874,
94.85532,
97.17808,
97.25629,
103.36186,
106.96647,
107.21461,
111.00887,
113.79269,
118.70564
),
intensity =
c(
8356.306,
7654.128,
9456.207,
8837.188,
8560.228,
8746.359,
8379.361,
169741.797,
7953.080,
8378.066
)
)
spectrum2 <- spectrum1
ms2_plot(spectrum1, spectrum2)
spectrum1 <- data.frame(
mz = c(
87.50874,
94.85532,
97.17808,
97.25629,
103.36186,
106.96647,
107.21461,
111.00887,
113.79269,
118.70564
),
intensity =
c(
8356.306,
7654.128,
9456.207,
8837.188,
8560.228,
8746.359,
8379.361,
169741.797,
7953.080,
8378.066
)
)
spectrum2 <- spectrum1
ms2_plot(spectrum1, spectrum2)
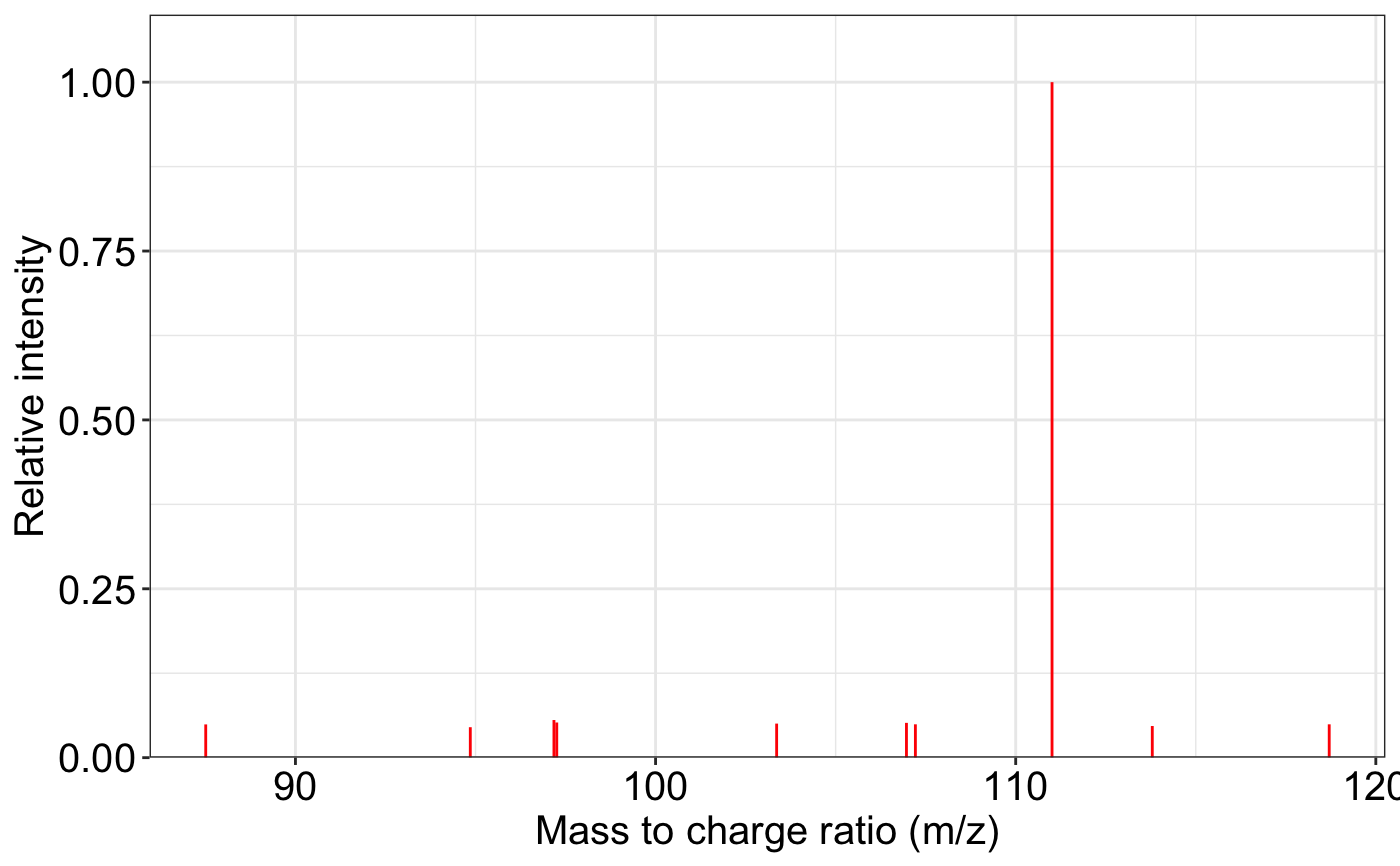 ms2_plot(spectrum1, spectrum2, interactive_plot = TRUE)
ms2_plot(spectrum1)
ms2_plot(spectrum1, spectrum2, interactive_plot = TRUE)
ms2_plot(spectrum1)
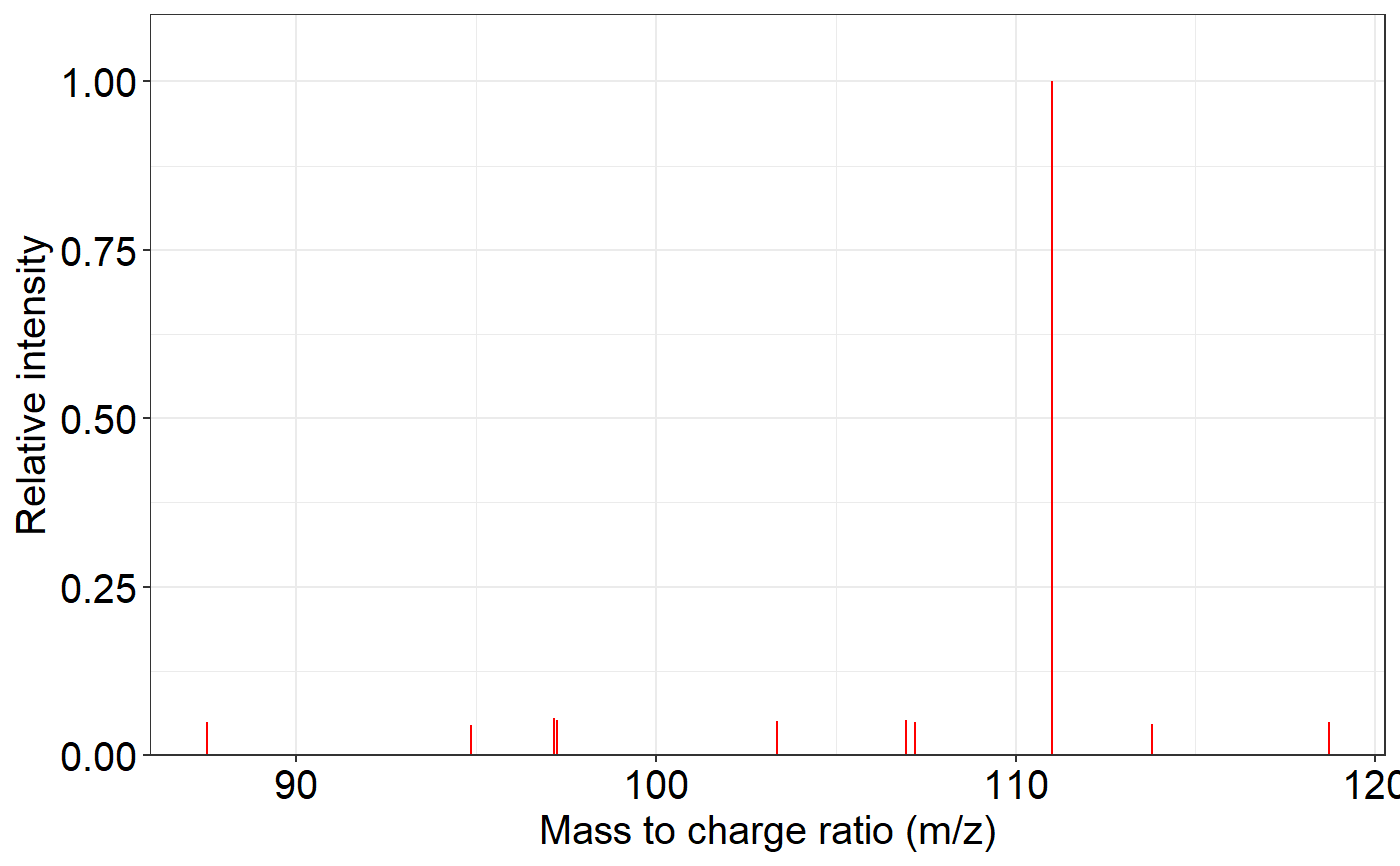 ms2_plot(spectrum1, interactive_plot = TRUE)
# Assuming `spec1_mat` and `spec2_mat` are matrices with MS2 spectra data
spec1_mat <- data.frame(
mz = c(
87.50874,
94.85532,
97.17808,
97.25629,
103.36186,
106.96647,
107.21461,
111.00887,
113.79269,
118.70564
),
intensity =
c(
8356.306,
7654.128,
9456.207,
8837.188,
8560.228,
8746.359,
8379.361,
169741.797,
7953.080,
8378.066
)
)
spec2_mat <- spec1_mat
ms2_plot(spec1_mat, spec2_mat)
ms2_plot(spectrum1, interactive_plot = TRUE)
# Assuming `spec1_mat` and `spec2_mat` are matrices with MS2 spectra data
spec1_mat <- data.frame(
mz = c(
87.50874,
94.85532,
97.17808,
97.25629,
103.36186,
106.96647,
107.21461,
111.00887,
113.79269,
118.70564
),
intensity =
c(
8356.306,
7654.128,
9456.207,
8837.188,
8560.228,
8746.359,
8379.361,
169741.797,
7953.080,
8378.066
)
)
spec2_mat <- spec1_mat
ms2_plot(spec1_mat, spec2_mat)