Introduction of masstools
Xiaotao Shen (https://www.shenxt.info/)
Created on 2022-02-18 and updated on 2023-10-01
Source:vignettes/masstools_introduction.Rmd
masstools_introduction.RmdBrief introduction
masstools is a package which contains multiple functions
for LC-MS metabolomics data processing and analysis. For example,
chemical formula operation, MS2 spectra matching. And
masstools is a part of tidymass project.
Chemical formula operation
You can use masstools to do the chemical formula
operation.
sum_formula(formula = "C9H11NO2", adduct = "M+H")
#> [1] "C9H12NO2"
sum_formula(formula = "C9H11NO2", adduct = "M+")
#> [1] "C9H11NO2"
sum_formula(formula = "C9H11NO2", adduct = "M+CH3COOH")
#> [1] "C11H15NO4"
sum_formula(formula = "C9H11", adduct = "M-H20")
#> [1] NAsplit_formula(formula = "C9H11NO2")
#> element.name number
#> 2 C 9
#> 3 H 11
#> 4 N 1
#> 5 O 2
split_formula(formula = "C2H4")
#> element.name number
#> 2 C 2
#> 3 H 4MS2 spectra operation
###remove the noisy peaks in one ms2 spectrum
exp.spectrum <- data.frame(mz = c(1:10, 1.0001),
intensity = c(1:10, 0.1))
ms2_plot(exp.spectrum)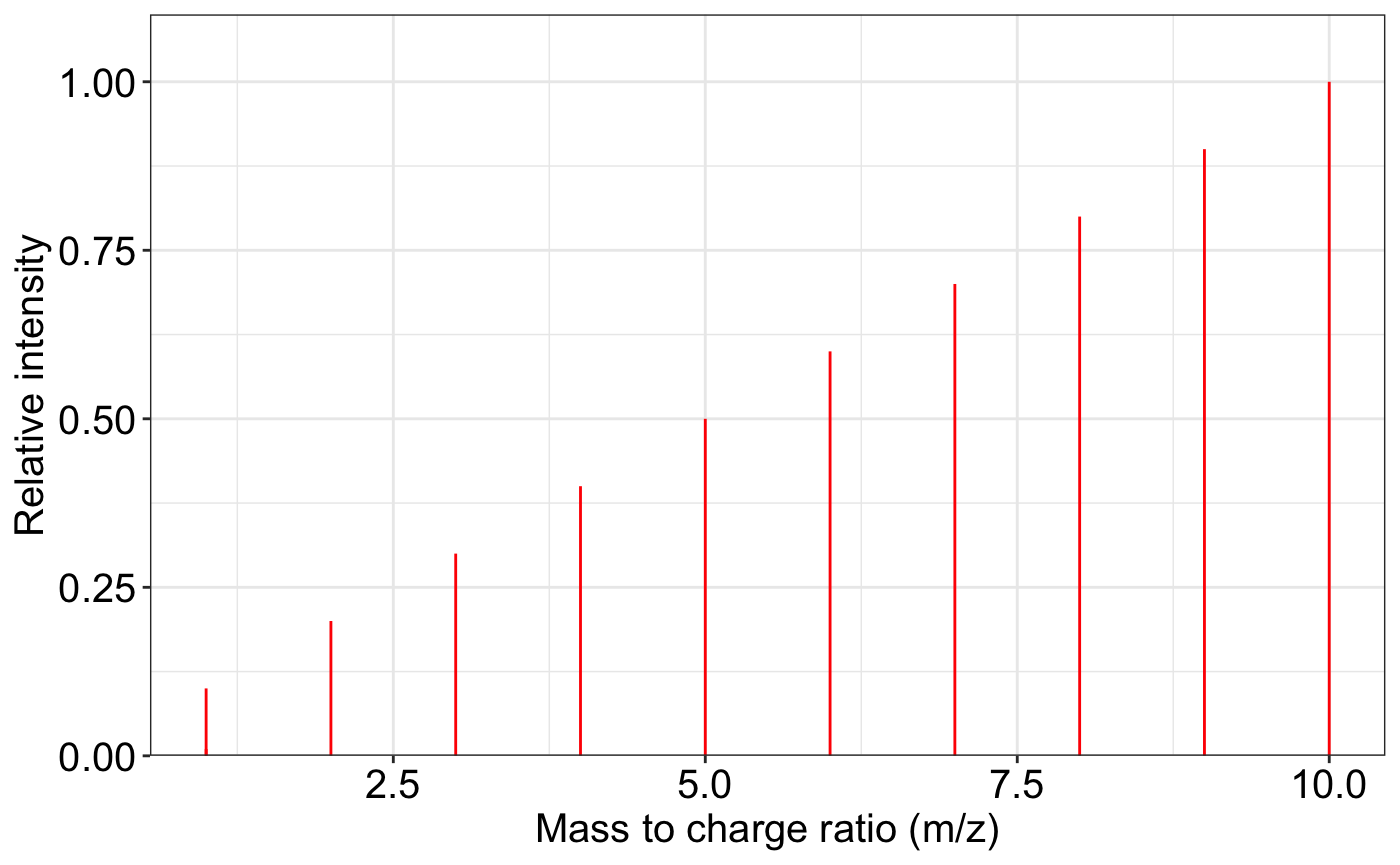
exp.spectrum2 = removeNoise(exp.spectrum)
ms2_plot(exp.spectrum, exp.spectrum2)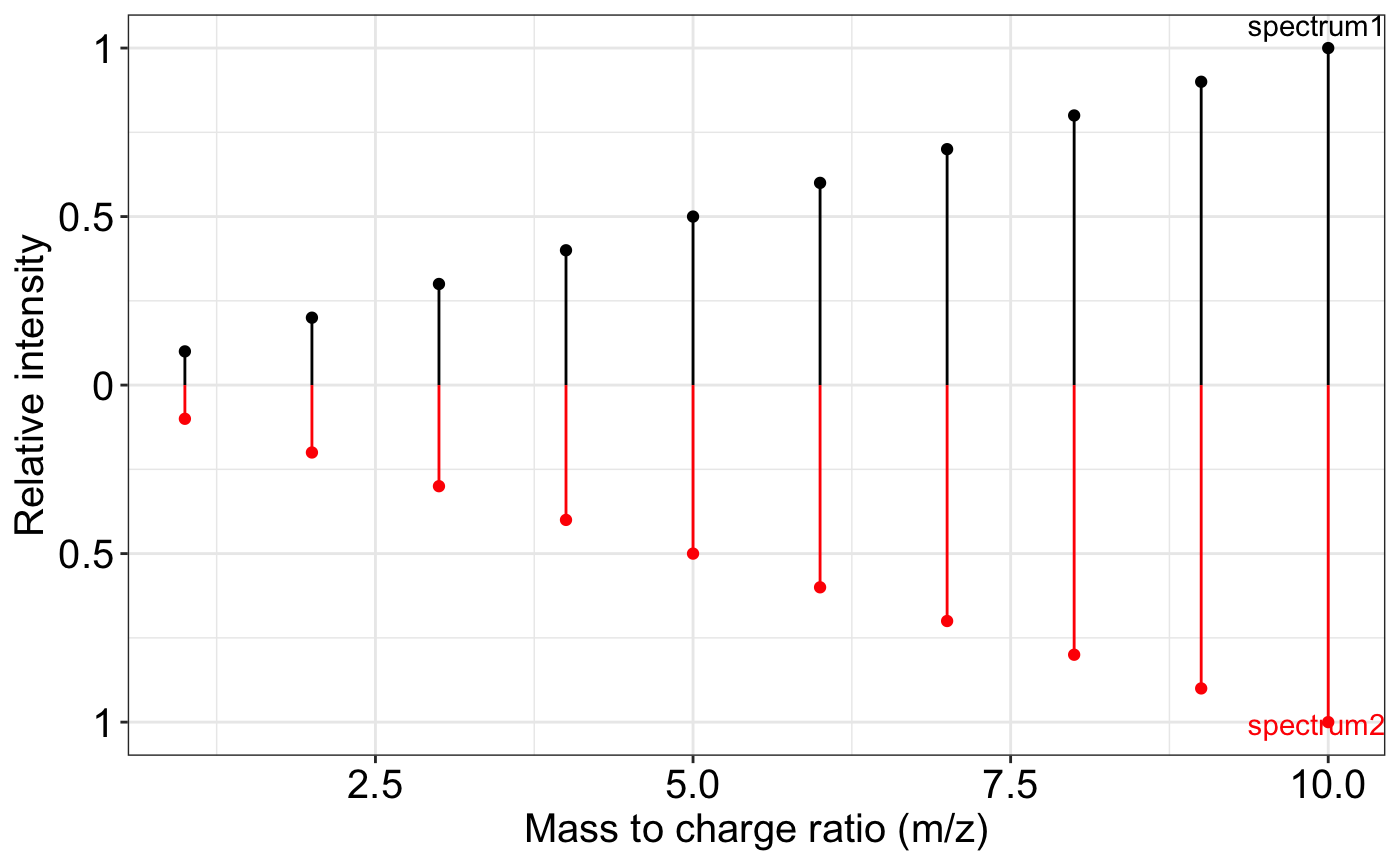
###match two spectra according to mz
exp.spectrum <- data.frame(mz = 1:10, intensity = 1:10)
lib.spectrum <- data.frame(mz = 1:10, intensity = 1:10)
ms2Match(exp.spectrum, lib.spectrum)
#> Lib.index Exp.index Lib.mz Lib.intensity Exp.mz Exp.intensity
#> 1 1 1 1 1 1 1
#> 2 2 2 2 2 2 2
#> 3 3 3 3 3 3 3
#> 4 4 4 4 4 4 4
#> 5 5 5 5 5 5 5
#> 6 6 6 6 6 6 6
#> 7 7 7 7 7 7 7
#> 8 8 8 8 8 8 8
#> 9 9 9 9 9 9 9
#> 10 10 10 10 10 10 10
## calculate the dot product of two matched intensity
getDP(exp.int = 1:10, lib.int = 1:10)
#> [1] 1
getDP(exp.int = 10:1, lib.int = 1:10)
#> [1] 0.379698
###matched two spectra and calculate dot product
exp.spectrum <- data.frame(mz = 1:10, intensity = 1:10)
lib.spectrum <- data.frame(mz = 1:10, intensity = 1:10)
getSpectraMatchScore(exp.spectrum, lib.spectrum)
#> [1] 1MS2 plot and MS2 matching plot.
spectrum1 <- data.frame(
mz = c(
87.50874,
94.85532,
97.17808,
97.25629,
103.36186,
106.96647,
107.21461,
111.00887,
113.79269,
118.70564
),
intensity =
c(
8356.306,
7654.128,
9456.207,
8837.188,
8560.228,
8746.359,
8379.361,
169741.797,
7953.080,
8378.066
)
)
spectrum2 <- spectrum1
ms2_plot(spectrum1, spectrum2)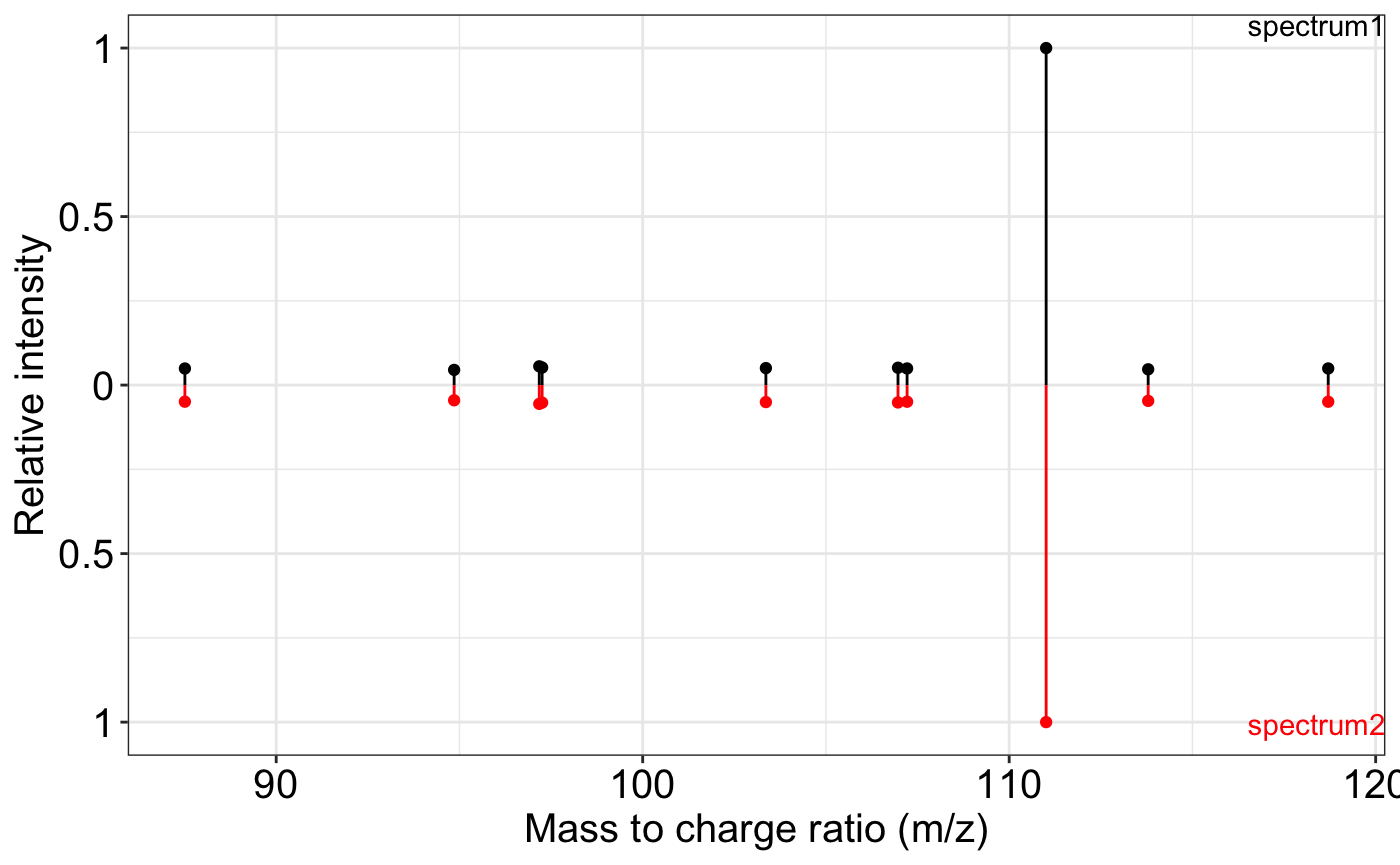
# ms2_plot(spectrum1, spectrum2, interactive_plot = TRUE)
ms2_plot(spectrum1)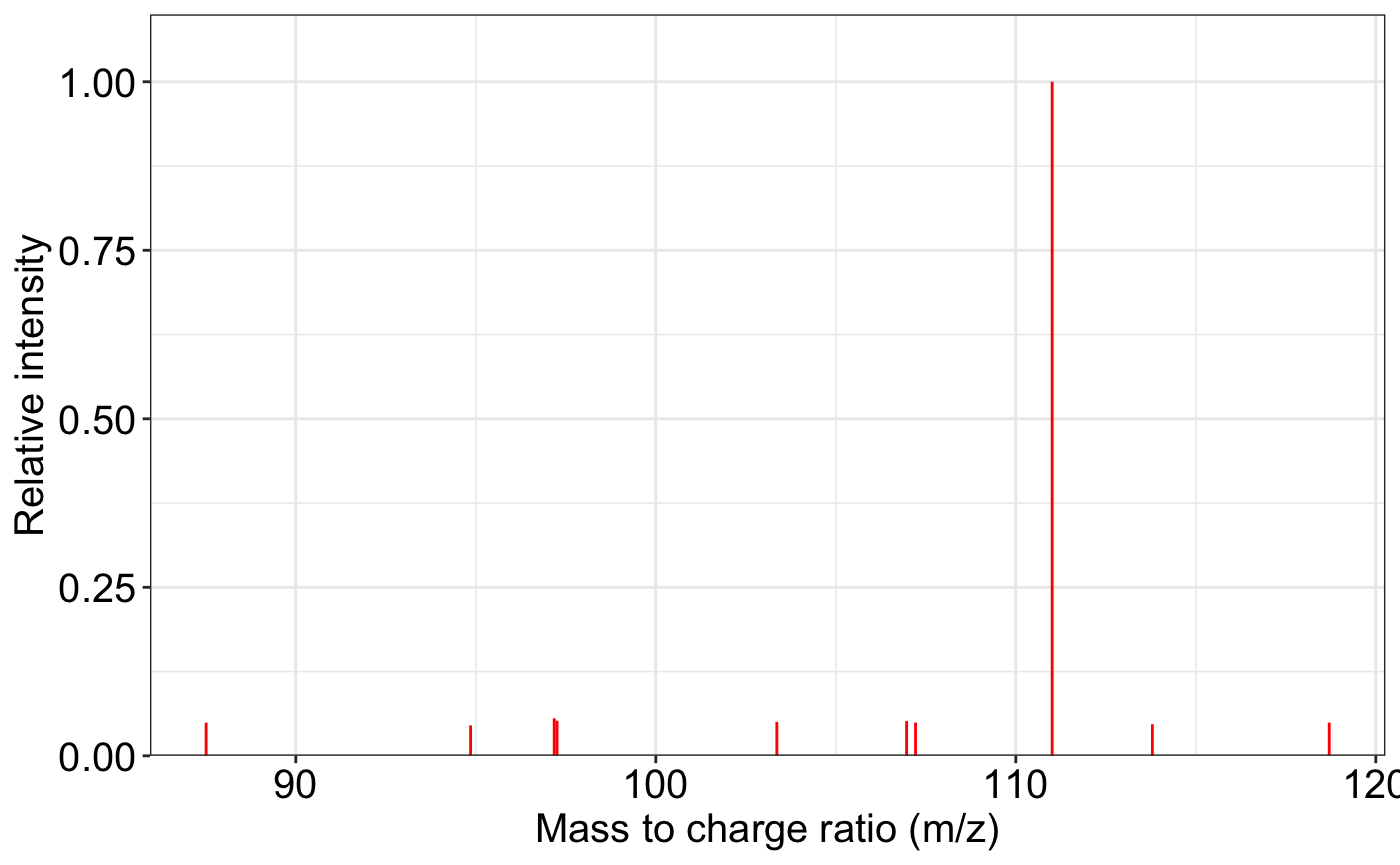
# ms2_plot(spectrum1, interactive_plot = TRUE)Match two feature tables
We can match two feature tables according to mz and retention time.
data1 <- data.frame(mz = 1:10, rt = 1:10)
data2 <- data.frame(mz = 1:10, rt = 1:10)
mz_rt_match(data1, data2, mz.tol = 10)
#> Index1 Index2 mz1 mz2 mz error rt1 rt2 rt error
#> 1 1 1 1 1 0 1 1 0
#> 2 2 2 2 2 0 2 2 0
#> 3 3 3 3 3 0 3 3 0
#> 4 4 4 4 4 0 4 4 0
#> 5 5 5 5 5 0 5 5 0
#> 6 6 6 6 6 0 6 6 0
#> 7 7 7 7 7 0 7 7 0
#> 8 8 8 8 8 0 8 8 0
#> 9 9 9 9 9 0 9 9 0
#> 10 10 10 10 10 0 10 10 0Compound ID converter
Two web tools are used for compound compound convert.
1. cts.fiehnlab
cts.fiehnlab is http://cts.fiehnlab.ucdavis.edu/service/convert. It support a lot of databases.
We can use the trans_id_database() to get the databases
that cts.fiehnlab.
database_name = trans_id_database(server = "cts.fiehnlab")head(database_name$From$From)
#> [1] "AAA Chemistry" "ABBLIS Chemicals" "Abbott Labs"
#> [4] "ABI Chem" "AbMole Bioscience" "Acesobio"head(database_name$To$From)
#> [1] "AAA Chemistry" "ABBLIS Chemicals" "Abbott Labs"
#> [4] "ABI Chem" "AbMole Bioscience" "Acesobio"We can see that it support a lot of (> 200) databases.
We can try the most common convert, from KEGG to HMDB.
trans_ID(
query = "C00001",
from = "KEGG",
to = "Human Metabolome Database",
top = 1,
server = "cts.fiehnlab"
)
#> KEGG Human Metabolome Database
#> 1 C00001 HMDB0002111Now, trans_ID doesn’t support verctor query. So you can
use the purrr::map() to achive this.
c("C00001", "C00001", "C00001") %>%
purrr::map(
.f = function(x) {
trans_ID(
query = x,
from = "KEGG",
to = "Human Metabolome Database",
top = 1,
server = "cts.fiehnlab"
)
}
) %>%
do.call(rbind, .) %>%
as.data.frame()
#> KEGG Human Metabolome Database
#> 1 C00001 HMDB0002111
#> 2 C00001 HMDB0002111
#> 3 C00001 HMDB00021112. chemspider
This is from https://www.chemspider.com/InChI.asmx.
We can use the trans_id_database() to get the databases
that chemspider
database_name2 = trans_id_database(server = "chemspider")database_name2$From
#> [1] "csid" "inchikey" "inchikey" "inchikey" "inchi" "inchi" "inchi"
#> [8] "inchi" "smiles"database_name2$To
#> [1] "mol" "csid" "inchi" "mol" "csid" "inchikey" "mol"
#> [8] "smiles" "inchi"This is very useful if you want to get the inchikey, inchi or smiles for one compound. But this web only support “ChemSpider ID” (csid), so we need use cts.fiehnlab convert to csid first.
trans_ID(
query = "C00001",
from = "KEGG",
to = "ChemSpider",
top = 1,
server = "cts.fiehnlab"
)
#> KEGG ChemSpider
#> 1 C00001 140526trans_ID(
query = "140526",
from = "csid",
to = "mol",
top = 1,
server = "chemspider"
)
#> [1] NAGet compound class based on classyfire
Refer this publication: https://jcheminf.biomedcentral.com/articles/10.1186/s13321-016-0174-y
result =
get_compound_class(
inchikey = "QZDWODWEESGPLC-UHFFFAOYSA-N",
server = "http://classyfire.wishartlab.com/entities/",
sleep = 5
)result
#> Kingdom : Organic compounds
#> └─Superclass : Organoheterocyclic compounds
#> └─Class : Pyridines and derivativesOther tools
Rename one vector with duplicated items.
name_duplicated(c("a", "a", "b", "c", "a", "b", "c", "a"))
#> [1] "a_1" "a_2" "b_1" "c_1" "a_3" "b_2" "c_2" "a_4"
name_duplicated(c(rep(1, 5), 2))
#> [1] "1_1" "1_2" "1_3" "1_4" "1_5" "2"
name_duplicated(1:5)
#> [1] 1 2 3 4 5Open the current working directory in R
####just open the current working directory
openwd()
###A new folder will be opened and pop upSet working directory in Windows
Copy the file path in File explorer in
Windows.
Then type in R:
setwd_win()Then paste the file path and type Enter.
Check the operate system
get_os()
#> [1] "osx"Check version of masstools
masstools_logo()
#> _______ _
#> |__ __| | |
#> _ __ ___ __ _ ___ ___| | ___ ___ | |___
#> | '_ ` _ \ / _` / __/ __| |/ _ \ / _ \| / __|
#> | | | | | | (_| \__ \__ \ | (_) | (_) | \__ \
#> |_| |_| |_|\__,_|___/___/_|\___/ \___/|_|___/
#>
#> Check conflicts of masstools
masstools_conflicts()
#> ── Conflicts ────────────────────────────────────────── masstools_conflicts() ──
#> ✖ methods::body<-() masks base::body<-()
#> ✖ tidyr::extract() masks magrittr::extract()
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ methods::kronecker() masks base::kronecker()
#> ✖ dplyr::lag() masks stats::lag()
#> ✖ purrr::set_names() masks magrittr::set_names()List all pacakges in masstools
masstools_packages()
#> [1] "dplyr" "remotes" "magrittr" "tibble" "tidyr"
#> [6] "stringr" "methods" "crayon" "cli" "purrr"
#> [11] "pbapply" "httr" "rvest" "xml2" "stats"
#> [16] "utils" "MSnbase" "ProtGenerics" "lifecycle" "ggplot2"
#> [21] "masstools"Session information
sessionInfo()
#> R version 4.3.0 (2023-04-21)
#> Platform: x86_64-apple-darwin20 (64-bit)
#> Running under: macOS 14.0
#>
#> Matrix products: default
#> BLAS: /Library/Frameworks/R.framework/Versions/4.3-x86_64/Resources/lib/libRblas.0.dylib
#> LAPACK: /Library/Frameworks/R.framework/Versions/4.3-x86_64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
#>
#> locale:
#> [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#>
#> time zone: America/Los_Angeles
#> tzcode source: internal
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] lubridate_1.9.2 forcats_1.0.0 stringr_1.5.0 purrr_1.0.1
#> [5] readr_2.1.4 tidyr_1.3.0 tibble_3.2.1 ggplot2_3.4.2
#> [9] tidyverse_2.0.0 dplyr_1.1.2 magrittr_2.0.3 masstools_1.0.13
#>
#> loaded via a namespace (and not attached):
#> [1] tidyselect_1.2.0 farver_2.1.1 fastmap_1.1.1
#> [4] XML_3.99-0.14 digest_0.6.31 timechange_0.2.0
#> [7] lifecycle_1.0.3 cluster_2.1.4 ProtGenerics_1.32.0
#> [10] compiler_4.3.0 rlang_1.1.1 sass_0.4.6
#> [13] tools_4.3.0 utf8_1.2.3 yaml_2.3.7
#> [16] knitr_1.43 labeling_0.4.2 curl_5.0.1
#> [19] xml2_1.3.4 plyr_1.8.8 BiocParallel_1.34.2
#> [22] withr_2.5.0 BiocGenerics_0.46.0 desc_1.4.2
#> [25] grid_4.3.0 stats4_4.3.0 preprocessCore_1.62.1
#> [28] fansi_1.0.4 colorspace_2.1-0 scales_1.2.1
#> [31] iterators_1.0.14 MASS_7.3-58.4 cli_3.6.1
#> [34] mzR_2.34.0 rmarkdown_2.22 crayon_1.5.2
#> [37] generics_0.1.3 remotes_2.4.2.1 rstudioapi_0.14
#> [40] httr_1.4.6 tzdb_0.4.0 ncdf4_1.21
#> [43] pbapply_1.7-0 cachem_1.0.8 affy_1.78.0
#> [46] zlibbioc_1.46.0 rvest_1.0.3 parallel_4.3.0
#> [49] impute_1.74.1 selectr_0.4-2 BiocManager_1.30.21
#> [52] vsn_3.68.0 vctrs_0.6.2 jsonlite_1.8.5
#> [55] IRanges_2.34.0 hms_1.1.3 S4Vectors_0.38.1
#> [58] MALDIquant_1.22.1 clue_0.3-64 foreach_1.5.2
#> [61] limma_3.56.2 jquerylib_0.1.4 affyio_1.70.0
#> [64] glue_1.6.2 MSnbase_2.26.0 pkgdown_2.0.7
#> [67] codetools_0.2-19 stringi_1.7.12 gtable_0.3.3
#> [70] mzID_1.38.0 munsell_0.5.0 pillar_1.9.0
#> [73] pcaMethods_1.92.0 htmltools_0.5.5 R6_2.5.1
#> [76] doParallel_1.0.17 rprojroot_2.0.3 evaluate_0.21
#> [79] lattice_0.21-8 Biobase_2.60.0 highr_0.10
#> [82] memoise_2.0.1 bslib_0.5.0 Rcpp_1.0.10
#> [85] xfun_0.39 MsCoreUtils_1.12.0 fs_1.6.2
#> [88] pkgconfig_2.0.3